Translation
Decoding the Transcript
After DNA is transcribed to RNA and the RNA is spliced, the next step in protein production is translation. The genetic code that cells use to translate RNA to protein is universally preserved across organisms. So you, your dog, and the lettuce in your salad all read and translate the genetic code the same. This helps scientists determine which changes in a gene have functional consequences by comparing transcripts from different species. Further, the universality of this code makes modeling genetic diseases in animals—such as grafting cancer onto a mouse, known as a xenograft—informative.
As a short aside, DNA and RNA use different coding alphabets: in a mRNA transcript, the DNA base thymine (T) is substituted by uracil (U). Thus, for all of the figures referencing mRNA, we will use U in the place of T.
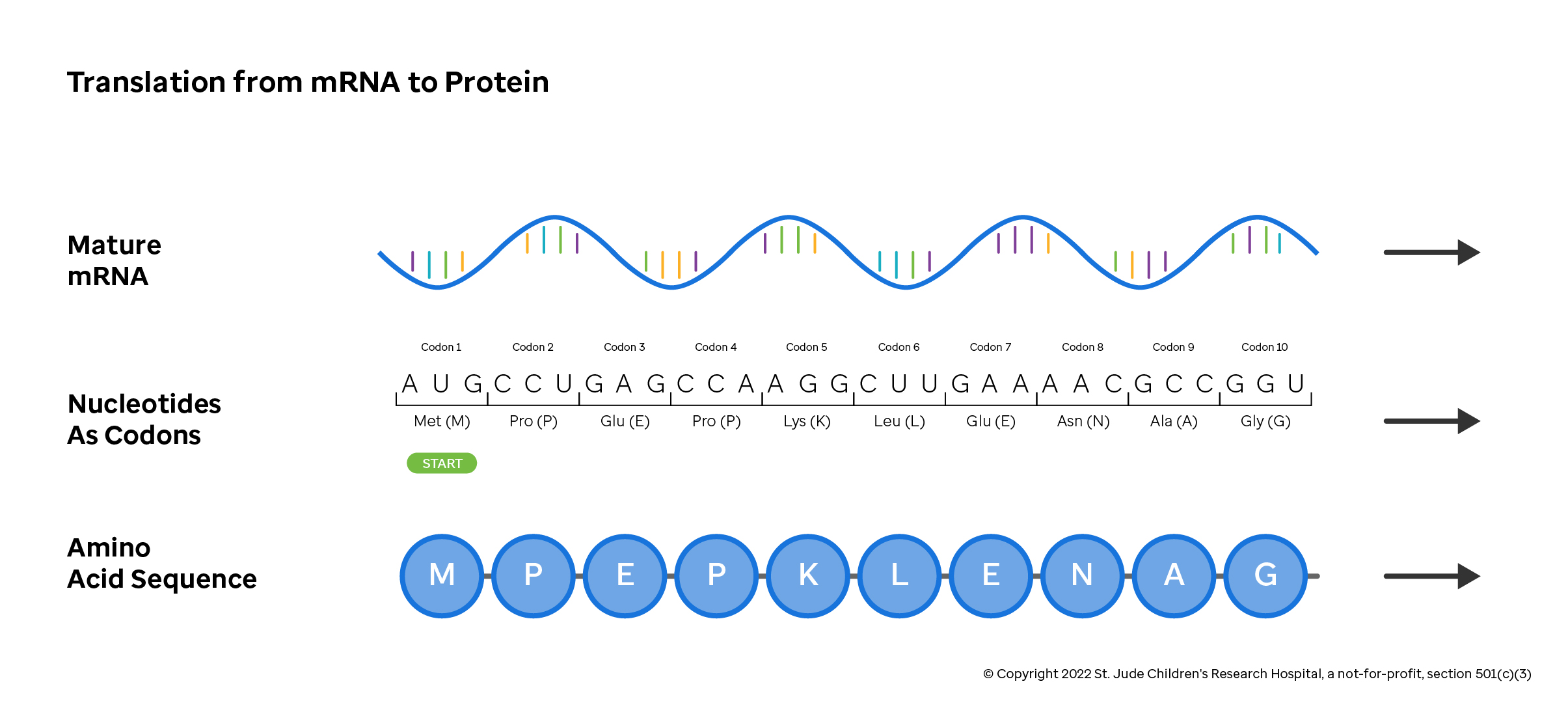
The mRNA transcript is encoded in three-letter nucleotide blocks known as codons. Codons specify that a particular protein building block, or amino acid, should be added to a growing protein chain. For example, the mRNA sequence above has "CCU" in the second codon position. This codon instructs the protein assembling machinery of the cell to add a Proline to the amino acid sequence at position two.
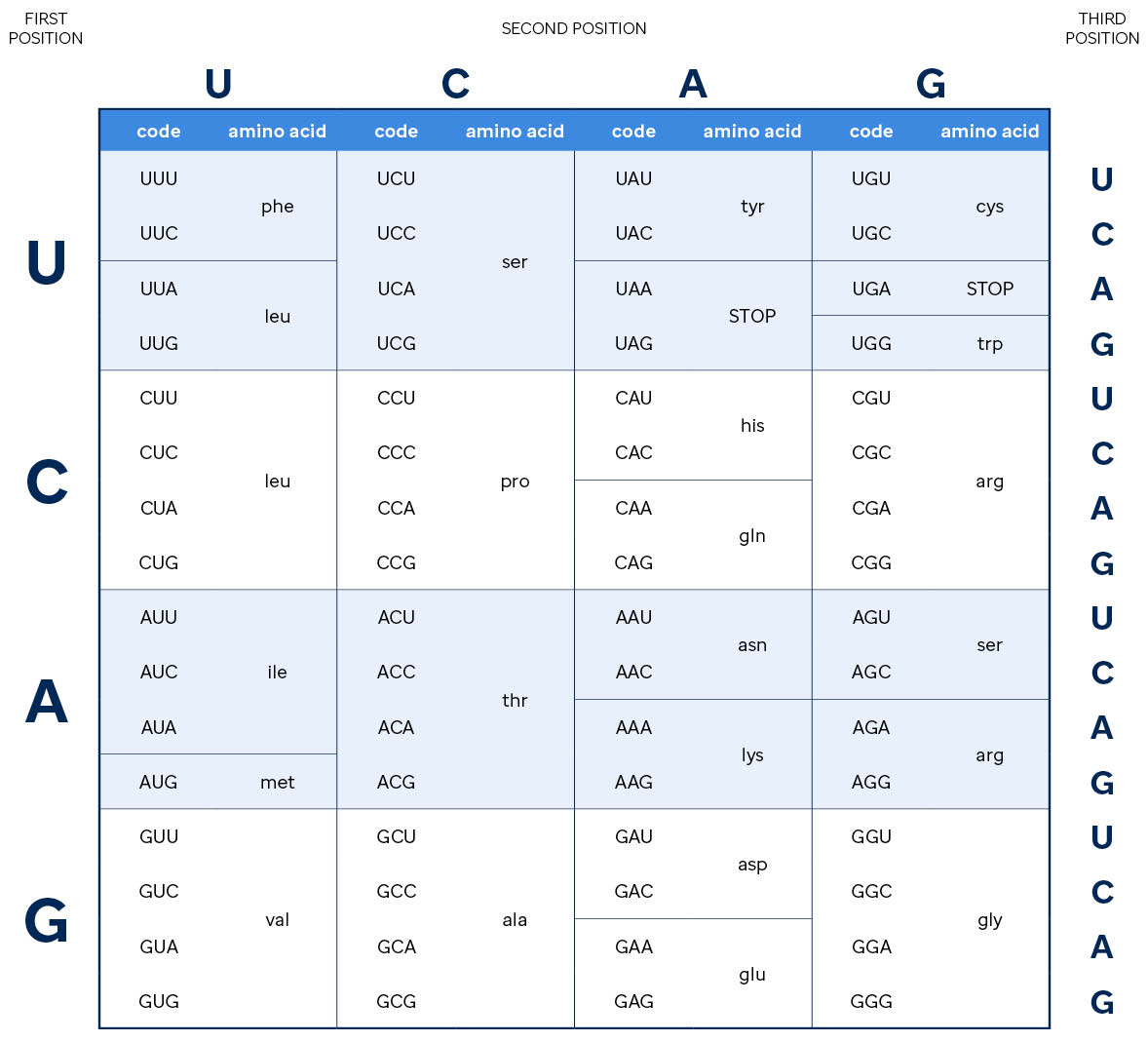
As shown in the amino acid lookup table below, other codons specify that translation should start or stop at a particular position. Further, multiple triplets translate to the same amino acid, some changes to the nucleotide sequence may not change which amino acid is added to the growing protein chain (e.g., if your triplet starts with "CG", the amino acid is always an arginine and the third nucleotide doesn't matter).
This class of variation, where the mRNA sequence changes but the resulting protein remains the same, is called a "silent mutation". There are many effects that changes in the mRNA sequence can have on the final protein byproduct, ranging from insignificant to deleterious. Knowledge of the codons and chemistry of proteins are used to predict the effect of a given variant on the resulting protein.
Reading Frames
Of course, any time you are decoding one alphabet into another, the accuracy of the process depends on your starting in the correct location. The three-letter code, and the resulting protein, will be completely different if translation starts on the first, second or third nucleotide.
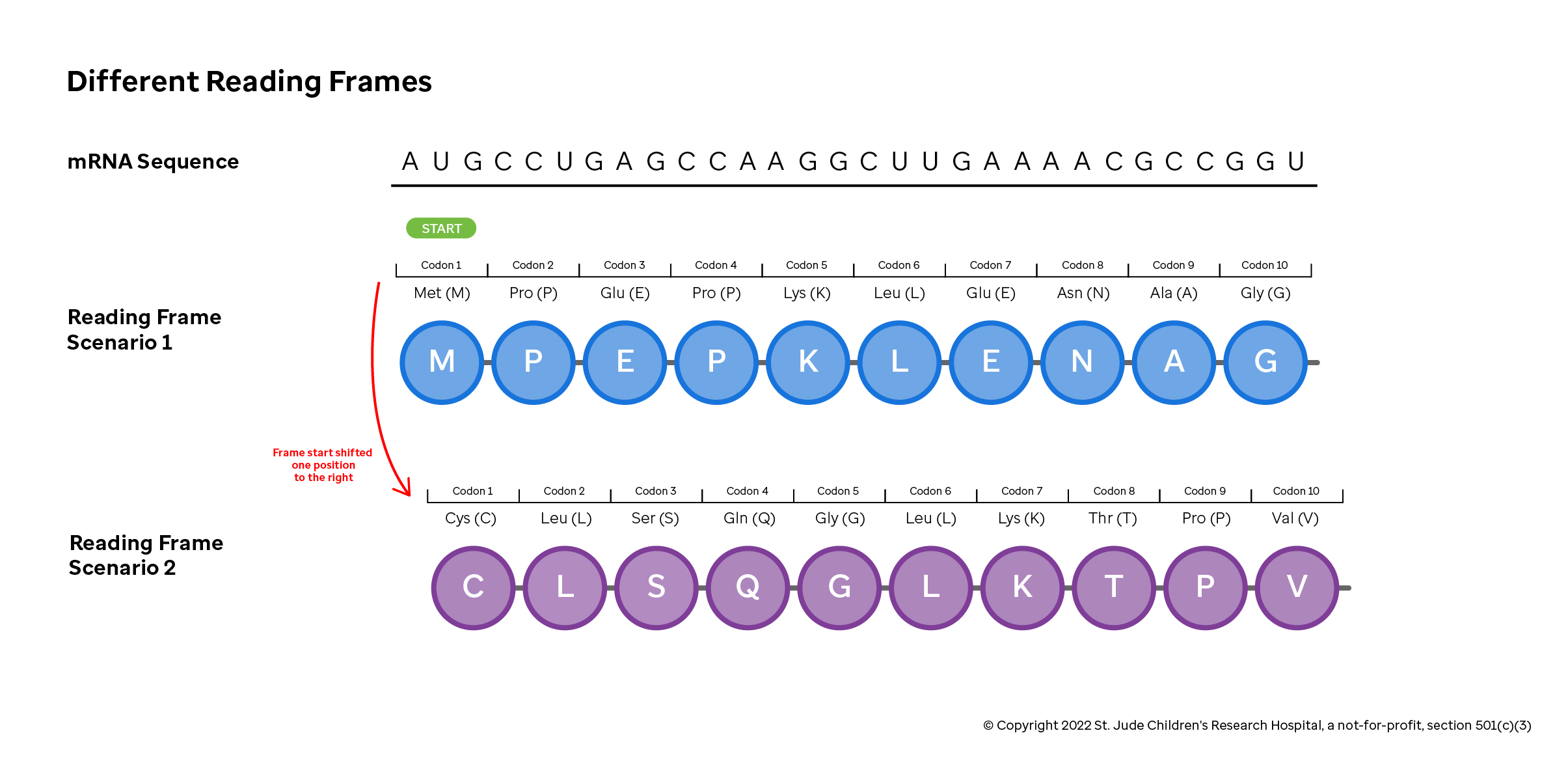
Consider the sequence in the figure above (a contrived example, but useful for our illustration here). If translation starts correctly at the first nucleotide, then the translation machinery correctly reads a start codon ("AUG" or Methionine), and then reads the following amino acids correctly. If the machinery were to start one nucleotide shifted to the right, it would read "UGC" (resulting in a Cysteine) and then a completely different protein would be made.
These different scenarios are referred to as reading frames, where the functional or open reading frame is one that begins with the start codon "AUG" (resulting in the Methionine). Changes to the sequence that starts, stops or moves a reading frame dramatically alters the protein product. Mutations that change which blocks of three nucleotides are used as codons are called frameshift mutations.