Types of Sequencing
Sequencing is a laboratory technique that identifies and determines the order of bases in a DNA molecule. Each sequencing type is best analyzed by a different computational method and is interpreted in a different way. The scientific goals of the research, cost, and practical limits determine which types of sequencing are chosen. For example, clinical sequencing seeks the time-sensitive detection of known mutations, while population studies focus on the discovery of new variants. Often, the type of sequencing ordered is a trade-off between cost and the breadth of information. This section discusses what scientists weigh when deciding which pieces of the genome/exome/transcriptome needs sequencing.
Common Sequence Applications
| Area of the genome | Abbreviations | Description |
|---|---|---|
| Whole Genome Sequencing | WGS | Characterizes the entire genome reading sequences from DNA |
| Whole Exome Sequencing | WES, WXS | Characterizes the exonic, or coding, regions of the genome reading sequences from DNA |
| Transcriptome Sequencing | WTS, RNA-seq | Characterizes the exonic regions that are actively expressed in the cell reading sequences from RNA |
| Targeted Sequencing | Validation Capture, Valcap | Characterizes a chosen region from DNA, can be a coding or non-coding region. The region can be coding or non-coding and is typically focused, high-depth sequencing for validating variants |
| ChIP seq | ChIP-seq | Characterizes the genomic locations where a protein interacts with DNA, both coding and non-coding |
| ATAC sequencing | ATAC-seq | Characterizes any sequence where DNA does not interact with proteins or is "open", both coding and non-coding |
| Hi-C | Hi-C | Characterizes the genomic locations where chromosomes come in close contact with each other. Typically used in characterizing 3D interactions of DNA |
| Single Cell Sequencing | scDNA-seq, scRNA-seq | DNA or RNA sequencing of individual cells, captures granularity at an individual cell level rather than a mixture of cells, can be coding or non-coding with lower coverage |
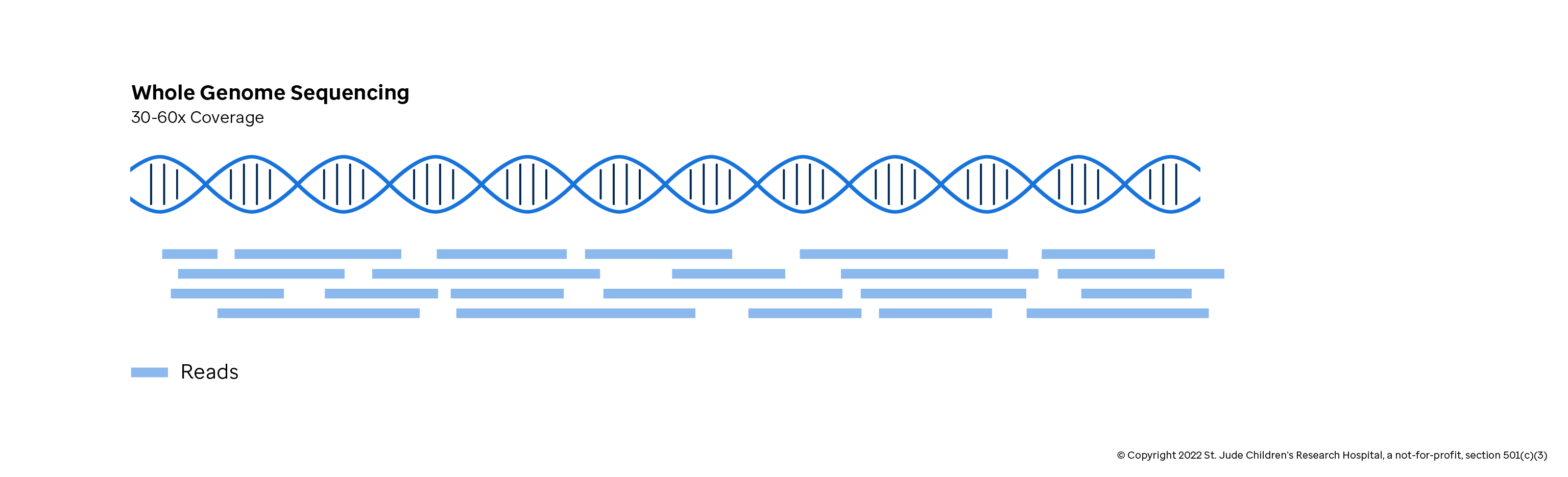
Whole Genome Sequencing
If cost was not an issue, whole genome sequencing (WGS) would be the standard approach for detecting variants. The advantage of WGS is a complete survey of the DNA, allowing the researcher to detect all the genomic variation for a given sample. WGS is needed when exploring how variations in non-coding regions influence the cells. Further, WGS is needed to detect the scope of variants that affect large regions of the chromosome (Structural Variation and Copy Number Variation, which are discussed in the "Genomic Variation" chapter). The disadvantage, other than cost, is that many variations are not relevant to disease or the research question and need to be carefully filtered out. WGS is used in basic exploratory research and increasingly elsewhere as costs drop.
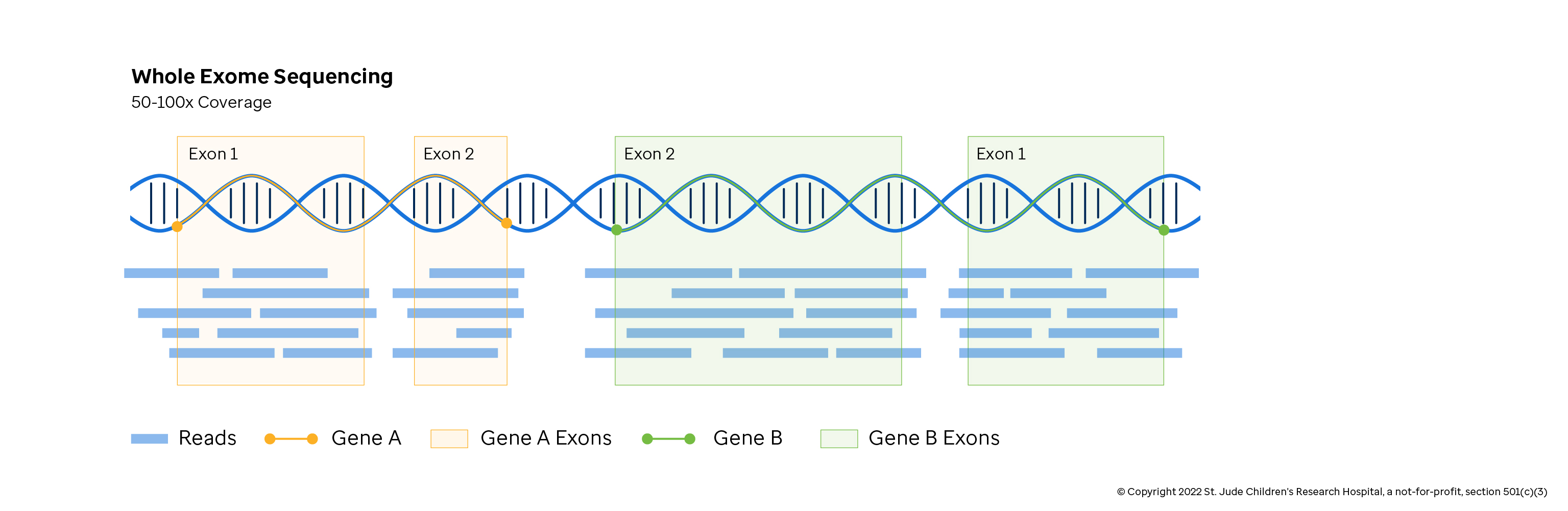
Whole Exome Sequencing
Whole exome sequencing (WES) captures the coding sequences of known genes from DNA and is a focused, cost-effective approach to characterizing what protein byproducts might be made. WES is commonly used to rapidly detect mutations in tumor samples so that treatments can be tailored to the individual. It is less commonly used in basic research due to its limited scope. The exceptions are large scale population studies with thousands of samples where whole genome approaches are too expensive.
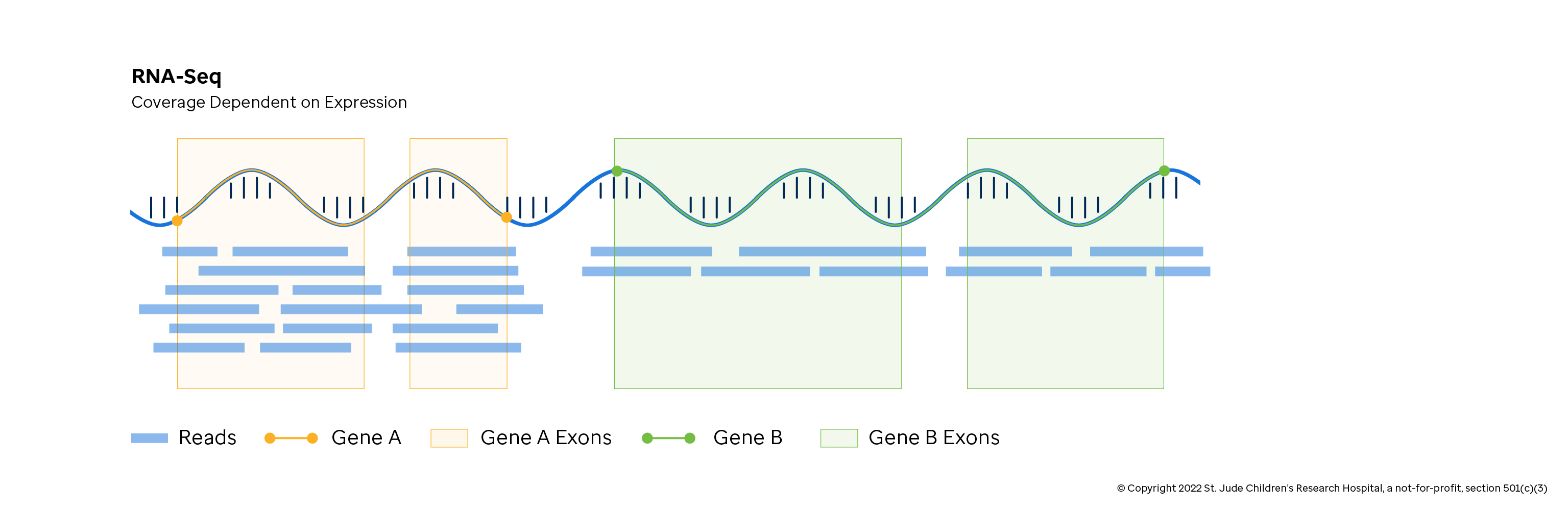
Whole Transcriptome Sequencing / RNA-seq
Whole transcriptome sequencing, or RNA-seq, captures the working recipes of genes as they float around a cell. This sequencing type gives the researcher a window into the mixture of proteins being created by the cell when the sample was collected. Mutations and natural variations, especially those which alter protein byproducts, can be detected using RNA-seq. Additionally, a common goal of RNA-seq is to quantify the transcripts of active genes to understand which genes are turned on and off in response to experimental treatments, disease or developmental changes in the cell.
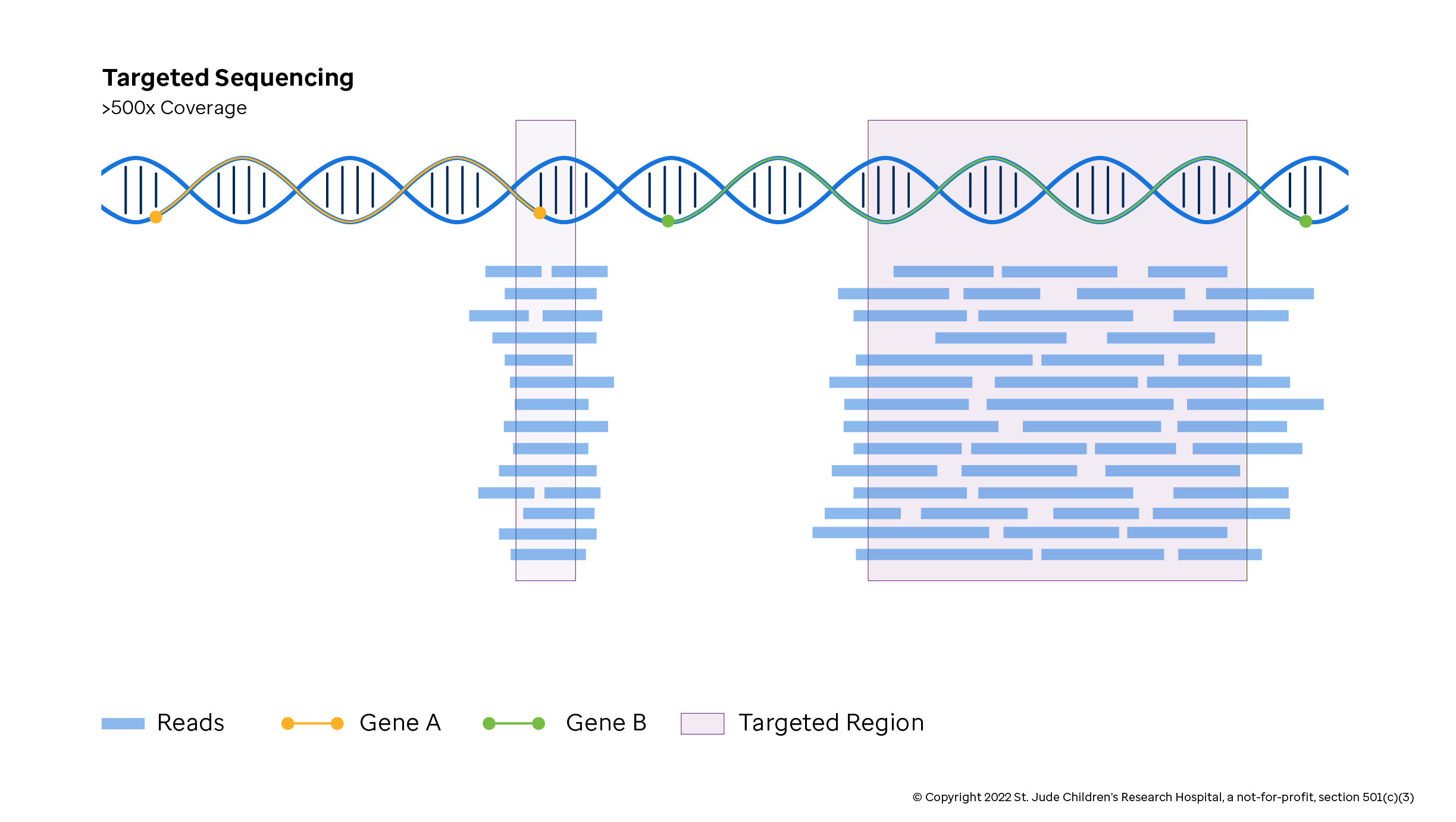
Targeted Sequencing
Targeted sequencing or validation capture sequencing (Valcap) captures preselected regions in the genome based on experimental design. Often targeted sequencing is used to capture a subset of important genes (called gene panels) in standardized tests when cost and time are limited. Alternatively, targeted sequencing is frequently used to gain greater insight into a region observed through a whole genome or whole exome study where there is less read depth. The method can be tailored for coding or non-coding variants.